Dawn Laney 1,2,3 and Jessica Dronen 3
Presented at 2021 National Society of Genetic Counselor (NSGC) Annual Conference
Background
Rare genetic conditions are notorious for being excluded from differential diagnosis of non-genetic specialists. This delay in diagnosis can lead to poorer health outcomes for patients and delay in treatment initiation. For example, Fabry disease has a median duration diagnostic delay of 10.5-21 years and studies have shown that early initiation with FDA approved therapies lead to optimal outcomes. A pilot study in two genetic conditions, Hereditary Angioedema (HAE) and Fabry disease (FD), was undertaken to determine if patients who may be at risk to have an underlying genetic condition can be flagged from de-identified medical records data fields.
Hypothesis
An automated severity scoring system can be used to screen patients at possible increased risk for Fabry disease (FD) or Hereditary Angioedema (HAE) with high sensitivity and specificity.
Methods
In this study an automated severity scoring system was created to score individuals as increased, possible, or no increased risk of FD or HAE. This scoring system was created with the consultation of experts in the field, as well as known clinical features of the conditions. This system was validated using published cases in the medical literature as well as anonymous patient data sets as positive controls and phenocopies as negative controls. Patients were determined to be at increased-risk if their raw scores were between 100 and 200 points. Patients were considered to be at high-risk if their raw scores were equal or greater than 200 points. An initial test run of the scoring system for HAE and FD was conducted on de-identified records of 298,288 patients. These records were obtained as part of a pilot study between ThinkGenetic, Inc. and Lafayette General Hospital (LGH). Records were requested for patients that presented to the facility with particular diagnosis codes, allergies, medications, and lab values over a period of approximately 6 months.
Discussion
The screening algorithms were found to have high sensitivities for FD and HAE as well as high specificities against the control groups. These results suggest that development, validation and implementation of automated severity scoring systems such as these can be useful screening tools to aid providers in identifying patients at risk for rare genetic conditions.
Results
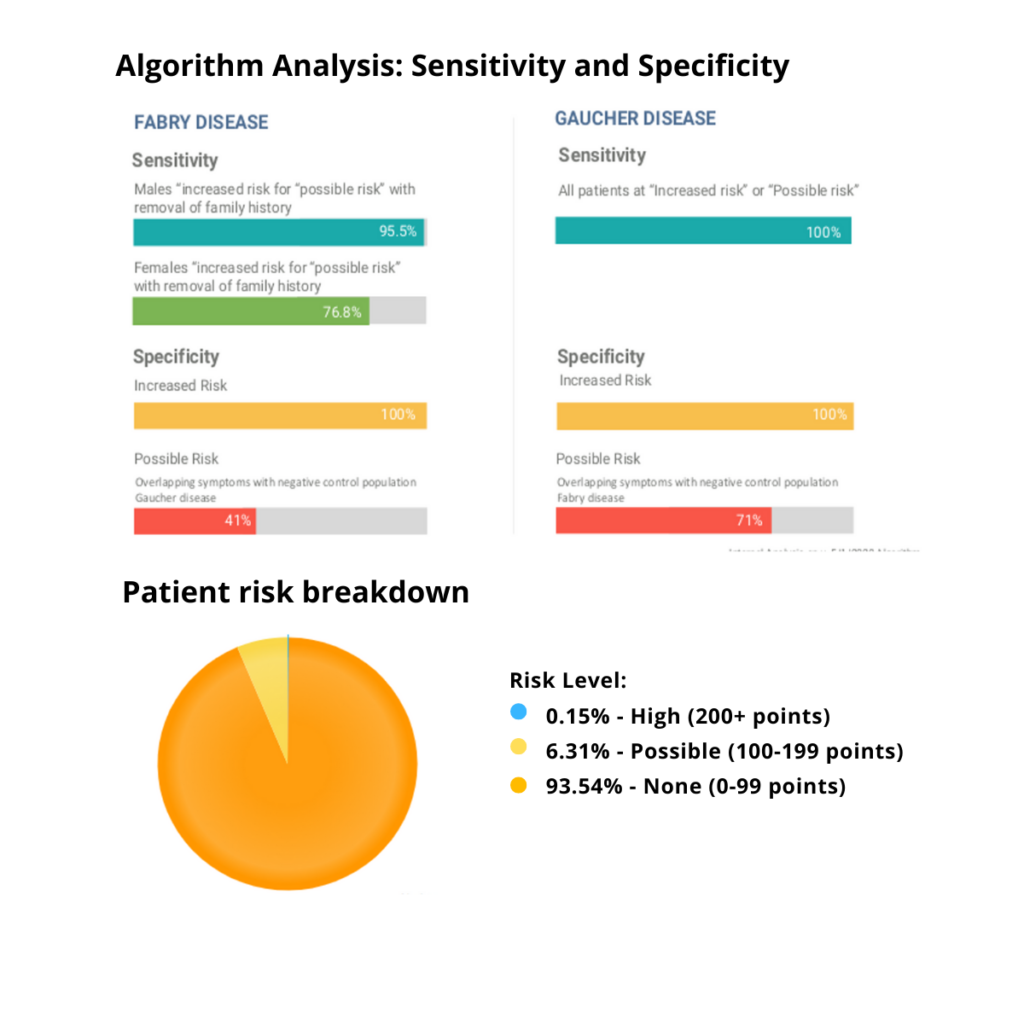
Validation: In FD, the scoring system was shown to have 95.5% sensitivity for males at increased or possible risk and a 76.8% sensitivity for females at increased or possible risk. There was 100% specificity for increased risk against the negative control population (Gaucher disease). In HAE there was a 98.6% sensitivity for individuals at increased risk or possible risk of HAE (including those with ACE inhibitor induced angioedema). There was 99% specificity against the negative control group (multifactorial edema) for increased risk.
Screening: The screening algorithm identified 297 patients as possible risk and 7 as increased risk for HAE. The algorithm identified 2 patients with a known family history of HAE, 2 individuals treated with HAE-specific medications, 54 individuals with low serum C4, and 241 individuals ACE inhibitor induced angioedema.
Future directions
Further studies are currently being conducted to implement additional screening algorithms. Additional studies for HAE and FD are also being conducted on a larger cohort of patients spanning a larger geographic area and longer period of time via partnership with a research network.
Limitations
Demographic information was not used to pre-screen patients included in this pilot study. All patients presenting at LGH during the study time period had equal opportunity to be included in analysis. However, this may not be a true representation of the area and may not generalize to other geographic locations. Due to the inability for follow-up in this pilot project we are not able to determine the clinical specificity and sensitivity of these algorithms at a particular facility.
Acknowledgements
This work was supported in part by an investigator initiated study grant funding from Takeda to the ThinkGenetic Foundation. We would also like to thank our colleagues at LGH, consulted experts in the field, our statistician colleagues, and our dedicated technical development team for their contributions to this project.
1 ThinkGenetic Foundation, Sudbury, MA, USA
2 Department of Human Genetics, Emory University, Atlanta, GA USA
3 ThinkGenetic, Inc. Lafayette, LA USA

