Dawn Laney, MS 1, 2 and Jessica Dronen, MS 3
2022 WORLD Symposium
Background
Outside the specialty of medical genetics, lysosomal storage disorders (LSDs) are rarely included in a patient’s differential diagnosis. This exclusion often leads to delays in diagnosis, disease specific monitoring, and treatment initiation. A pilot study in ten LSDs (Fabry disease, Gaucher disease, late-onset Pompe disease, Niemann Pick type B, Mucopolysaccharidosis types I/II/IV/VI/VII, and Farber disease) was undertaken to determine if at-risk patients can be flagged from de-identified electronic medical record data fields.
Methods
Disease-specific automated severity scoring systems were created to score individuals as increased, possible, or no increased risk. The system was initially validated using published cases and anonymous patient data sets as positive controls. Phenocopies for each individual LSD were used as negative controls.
Results
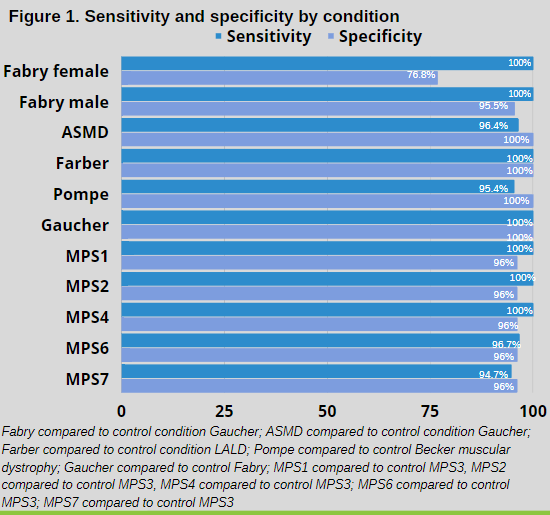
In FD, the scoring system was shown to have 95.5% sensitivity for males at increased or possible risk and a 76.8% sensitivity for females at increased or possible risk. The specificity approached 100% for increased risk against the negative control populations. In the other conditions, the sensitivity and specificity for individuals at increased risk or possible risk approached 95-100% (See Figure 1 for condition-specific sensitivities and specificities).
Future Directions
Following validation, test runs of the scoring systems were conducted on de-identified electronic health records of over 100,000 real-world patients. Further refinement of the screening algorithms is being completed to optimize performance in live electronic health records. Further studies are currently being conducted to implement screening algorithms to identify at-risk patients in electronic data fields from several large medical centers across the United States.
Acknowledgements
This work was supported in partnership with the ThinkGenetic Foundation. We would also like to thank our colleagues at Emory University, consulted experts in the field, our statistician colleagues, and our dedicated technical development team for their contributions to this project.
1 Emory University
2 ThinkGenetic Foundation
3 ThinkGenetic, Inc.

